Are Golgi Bodies In Plant And Animal Cells
| Jail cell biology | |
|---|---|
| Fauna cell diagram | |
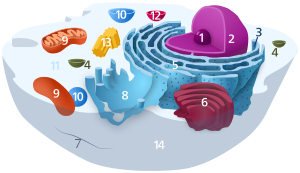 Components of a typical creature jail cell:
|

Micrograph of Golgi appliance, visible equally a stack of semicircular black rings about the lesser. Numerous round vesicles can be seen in proximity to the organelle.
The Golgi appliance (), also known equally the Golgi circuitous, Golgi torso, or simply the Golgi, is an organelle found in most eukaryotic cells.[1] Part of the endomembrane system in the cytoplasm, it packages proteins into membrane-jump vesicles inside the jail cell before the vesicles are sent to their destination. It resides at the intersection of the secretory, lysosomal, and endocytic pathways. Information technology is of detail importance in processing proteins for secretion, containing a gear up of glycosylation enzymes that adhere diverse saccharide monomers to proteins equally the proteins motility through the appliance.
It was identified in 1897 past the Italian scientist Camillo Golgi and was named later him in 1898.[2]
Discovery
Owing to its big size and distinctive structure, the Golgi apparatus was ane of the first organelles to be discovered and observed in detail. It was discovered in 1898 by Italian physician Camillo Golgi during an investigation of the nervous system.[iii] [2] Later on first observing information technology under his microscope, he termed the construction as apparato reticolare interno ("internal reticular appliance"). Some doubted the discovery at outset, arguing that the appearance of the structure was only an optical illusion created by the observation technique used past Golgi. With the development of mod microscopes in the twentieth century, the discovery was confirmed.[4] Early on references to the Golgi appliance referred to it by various names including the "Golgi–Holmgren apparatus", "Golgi–Holmgren ducts", and "Golgi–Kopsch appliance".[2] The term "Golgi apparatus" was used in 1910 and showtime appeared in the scientific literature in 1913, while "Golgi complex" was introduced in 1956.[2]
Subcellular localization
The subcellular localization of the Golgi appliance varies amidst eukaryotes. In mammals, a single Golgi appliance is usually located near the cell nucleus, shut to the centrosome. Tubular connections are responsible for linking the stacks together. Localization and tubular connections of the Golgi appliance are dependent on microtubules. In experiments information technology is seen that as microtubules are depolymerized the Golgi apparatuses lose common connections and go private stacks throughout the cytoplasm.[five] In yeast, multiple Golgi apparatuses are scattered throughout the cytoplasm (as observed in Saccharomyces cerevisiae). In plants, Golgi stacks are non concentrated at the centrosomal region and exercise not form Golgi ribbons.[6] Organization of the institute Golgi depends on actin cables and not microtubules.[vi] The common feature amid Golgi is that they are adjacent to endoplasmic reticulum (ER) exit sites.[7]
Structure

3D rendering of Golgi appliance

Diagram of a single "stack" of Golgi
In most eukaryotes, the Golgi apparatus is made up of a series of compartments and is a collection of fused, flattened membrane-enclosed disks known equally cisternae (singular: cisterna, also called "dictyosomes"), originating from vesicular clusters that bud off the endoplasmic reticulum. A mammalian jail cell typically contains forty to 100 stacks of cisternae.[8] Between four and eight cisternae are usually present in a stack; notwithstanding, in some protists as many as threescore cisternae have been observed.[4] This collection of cisternae is broken down into cis, medial, and trans compartments, making up two main networks: the cis Golgi network (CGN) and the trans Golgi network (TGN). The CGN is the first cisternal structure, and the TGN is the final, from which proteins are packaged into vesicles destined to lysosomes, secretory vesicles, or the cell surface. The TGN is usually positioned adjacent to the stack, merely tin too be divide from it. The TGN may act every bit an early endosome in yeast and plants.[half dozen] [nine]
There are structural and organizational differences in the Golgi apparatus among eukaryotes. In some yeasts, Golgi stacking is not observed. Pichia pastoris does take stacked Golgi, while Saccharomyces cerevisiae does not.[6] In plants, the individual stacks of the Golgi apparatus seem to operate independently.[6]
The Golgi apparatus tends to be larger and more than numerous in cells that synthesize and secrete large amounts of substances; for example, the antibiotic-secreting plasma B cells of the immune system accept prominent Golgi complexes.
In all eukaryotes, each cisternal stack has a cis entry face up and a trans exit face. These faces are characterized by unique morphology and biochemistry.[10] Within individual stacks are assortments of enzymes responsible for selectively modifying protein cargo. These modifications influence the fate of the protein. The compartmentalization of the Golgi apparatus is advantageous for separating enzymes, thereby maintaining consecutive and selective processing steps: enzymes catalyzing early modifications are gathered in the cis face up cisternae, and enzymes catalyzing later modifications are found in trans face cisternae of the Golgi stacks.[v] [ten]
Office

The Golgi apparatus (salmon pinkish) in context of the secretory pathway
The Golgi apparatus is a major collection and acceleration station of protein products received from the endoplasmic reticulum (ER). Proteins synthesized in the ER are packaged into vesicles, which so fuse with the Golgi apparatus. These cargo proteins are modified and destined for secretion via exocytosis or for apply in the cell. In this respect, the Golgi can be thought of as like to a post office: it packages and labels items which it and so sends to dissimilar parts of the prison cell or to the extracellular space. The Golgi apparatus is also involved in lipid ship and lysosome formation.[eleven]
The structure and part of the Golgi apparatus are intimately linked. Individual stacks have unlike assortments of enzymes, allowing for progressive processing of cargo proteins as they travel from the cisternae to the trans Golgi face.[5] [10] Enzymatic reactions within the Golgi stacks occur exclusively about its membrane surfaces, where enzymes are anchored. This feature is in contrast to the ER, which has soluble proteins and enzymes in its lumen. Much of the enzymatic processing is post-translational modification of proteins. For example, phosphorylation of oligosaccharides on lysosomal proteins occurs in the early CGN.[5] Cis cisterna are associated with the removal of mannose residues.[5] [ten] Removal of mannose residues and addition of N-acetylglucosamine occur in medial cisternae.[five] Addition of galactose and sialic acid occurs in the trans cisternae.[5] Sulfation of tyrosines and carbohydrates occurs within the TGN.[five] Other general mail-translational modifications of proteins include the addition of carbohydrates (glycosylation)[12] and phosphates (phosphorylation). Protein modifications may grade a signal sequence that determines the concluding destination of the protein. For example, the Golgi appliance adds a mannose-6-phosphate label to proteins destined for lysosomes. Another of import function of the Golgi apparatus is in the formation of proteoglycans. Enzymes in the Golgi append proteins to glycosaminoglycans, thus creating proteoglycans.[13] Glycosaminoglycans are long unbranched polysaccharide molecules nowadays in the extracellular matrix of animals.
Vesicular transport

The vesicles that exit the rough endoplasmic reticulum are transported to the cis confront of the Golgi appliance, where they fuse with the Golgi membrane and empty their contents into the lumen. Once inside the lumen, the molecules are modified, then sorted for transport to their adjacent destinations.
Those proteins destined for areas of the cell other than either the endoplasmic reticulum or the Golgi apparatus are moved through the Golgi cisternae towards the trans face, to a complex network of membranes and associated vesicles known equally the trans-Golgi network (TGN). This area of the Golgi is the point at which proteins are sorted and shipped to their intended destinations by their placement into one of at least three different types of vesicles, depending upon the signal sequence they carry.
| Types | Description | Example |
|---|---|---|
| Exocytotic vesicles (constitutive) | Vesicle contains proteins destined for extracellular release. Later on packaging, the vesicles bud off and immediately move towards the plasma membrane, where they fuse and release the contents into the extracellular space in a procedure known as constitutive secretion. | Antibody release past activated plasma B cells |
| Secretory vesicles (regulated) | Vesicles contain proteins destined for extracellular release. Later packaging, the vesicles bud off and are stored in the cell until a point is given for their release. When the advisable signal is received they move toward the membrane and fuse to release their contents. This process is known equally regulated secretion. | Neurotransmitter release from neurons |
| Lysosomal vesicles | Vesicles comprise proteins and ribosomes destined for the lysosome, a degradative organelle containing many acid hydrolases, or to lysosome-like storage organelles. These proteins include both digestive enzymes and membrane proteins. The vesicle first fuses with the late endosome, and the contents are and so transferred to the lysosome via unknown mechanisms. | Digestive proteases destined for the lysosome |
Current models of vesicular transport and trafficking
Model 1: Anterograde vesicular transport betwixt stable compartments
- In this model, the Golgi is viewed equally a set of stable compartments that work together. Each compartment has a unique collection of enzymes that piece of work to alter protein cargo. Proteins are delivered from the ER to the cis confront using COPII-coated vesicles. Cargo then progress toward the trans face in COPI-coated vesicles. This model proposes that COPI vesicles move in two directions: anterograde vesicles conduct secretory proteins, while retrograde vesicles recycle Golgi-specific trafficking proteins.[14]
- Strengths: The model explains observations of compartments, polarized distribution of enzymes, and waves of moving vesicles. It also attempts to explicate how Golgi-specific enzymes are recycled.[xiv]
- Weaknesses: Since the corporeality of COPI vesicles varies drastically among types of cells, this model cannot hands explain loftier trafficking activeness inside the Golgi for both small and large cargoes. Additionally, there is no disarming evidence that COPI vesicles motility in both the anterograde and retrograde directions.[14]
- This model was widely accepted from the early 1980s until the late 1990s.[14]
Model 2: Cisternal progression/maturation
- In this model, the fusion of COPII vesicles from the ER begins the formation of the first cis-cisterna of the Golgi stack, which progresses afterward to become mature TGN cisternae. Once matured, the TGN cisternae dissolve to get secretory vesicles. While this progression occurs, COPI vesicles continually recycle Golgi-specific proteins by delivery from older to younger cisternae. Different recycling patterns may account for the differing biochemistry throughout the Golgi stack. Thus, the compartments within the Golgi are seen as detached kinetic stages of the maturing Golgi appliance.[xiv]
- Strengths: The model addresses the beingness of Golgi compartments, every bit well as differing biochemistry within the cisternae, send of large proteins, transient formation and disintegration of the cisternae, and retrograde mobility of native Golgi proteins, and it can account for the variability seen in the structures of the Golgi.[fourteen]
- Weaknesses: This model cannot easily explain the observation of fused Golgi networks, tubular connections among cisternae, and differing kinetics of secretory cargo leave.[fourteen]
Model 3: Cisternal progression/maturation with heterotypic tubular transport
- This model is an extension of the cisternal progression/maturation model. It incorporates the existence of tubular connections amongst the cisternae that form the Golgi ribbon, in which cisternae within a stack are linked. This model posits that the tubules are important for bidirectional traffic in the ER-Golgi system: they let for fast anterograde traffic of pocket-sized cargo and/or the retrograde traffic of native Golgi proteins.[14] [xv]
- Strengths: This model encompasses the strengths of the cisternal progression/maturation model that also explains rapid trafficking of cargo, and how native Golgi proteins can recycle independently of COPI vesicles.[14]
- Weaknesses: This model cannot explain the transport kinetics of large poly peptide cargo, such as collagen. Additionally, tubular connections are not prevalent in plant cells. The roles that these connections have can be attributed to a cell-specific specialization rather than a universal trait. If the membranes are continuous, that suggests the beingness of mechanisms that preserve the unique biochemical gradients observed throughout the Golgi apparatus.[xiv]
Model 4: Rapid partitioning in a mixed Golgi
- This rapid sectionalisation model is the most drastic alteration of the traditional vesicular trafficking point of view. Proponents of this model hypothesize that the Golgi works as a single unit of measurement, containing domains that part separately in the processing and export of poly peptide cargo. Cargo from the ER movement between these ii domains, and randomly leave from whatever level of the Golgi to their final location. This model is supported by the observation that cargo exits the Golgi in a pattern all-time described by exponential kinetics. The being of domains is supported by fluorescence microscopy information.[14]
- Strengths: Notably, this model explains the exponential kinetics of cargo go out of both large and small proteins, whereas other models cannot.[14]
- Weaknesses: This model cannot explicate the transport kinetics of large protein cargo, such as collagen. This model falls short on explaining the observation of discrete compartments and polarized biochemistry of the Golgi cisternae. Information technology besides does not explain formation and disintegration of the Golgi network, nor the function of COPI vesicles.[14]
Model 5: Stable compartments as cisternal model progenitors
- This is the about contempo model. In this model, the Golgi is seen as a drove of stable compartments defined by Rab (One thousand-protein) GTPases.[14]
- Strengths: This model is consistent with numerous observations and encompasses some of the strengths of the cisternal progression/maturation model. Additionally, what is known of the Rab GTPase roles in mammalian endosomes can assistance predict putative roles inside the Golgi. This model is unique in that it tin can explain the observation of "megavesicle" transport intermediates.[14]
- Weaknesses: This model does not explicate morphological variations in the Golgi apparatus, nor define a office for COPI vesicles. This model does not apply well for plants, algae, and fungi in which individual Golgi stacks are observed (transfer of domains between stacks is not likely). Additionally, megavesicles are non established to be intra-Golgi transporters.[14]
Though there are multiple models that attempt to explain vesicular traffic throughout the Golgi, no private model can independently explain all observations of the Golgi appliance. Currently, the cisternal progression/maturation model is the well-nigh accustomed among scientists, accommodating many observations across eukaryotes. The other models are still of import in framing questions and guiding future experimentation. Among the fundamental unanswered questions are the directionality of COPI vesicles and role of Rab GTPases in modulating poly peptide cargo traffic.[fourteen]
Brefeldin A
Brefeldin A (BFA) is a fungal metabolite used experimentally to disrupt the secretion pathway as a method of testing Golgi function.[16] BFA blocks the activation of some ADP-ribosylation factors (ARFs).[17] ARFs are small GTPases which regulate vesicular trafficking through the binding of COPs to endosomes and the Golgi.[17] BFA inhibits the function of several guanine nucleotide substitution factors (GEFs) that mediate GTP-binding of ARFs.[17] Handling of cells with BFA thus disrupts the secretion pathway, promoting disassembly of the Golgi apparatus and distributing Golgi proteins to the endosomes and ER.[16] [17]
Gallery
-
Yeast Golgi dynamics. Green labels early Golgi, blood-red labels tardily Golgi.[18]
-

Two Golgi stacks connected as a ribbon in a mouse prison cell. Taken from the movie.
References
- ^ Pavelk Thousand, Mironov AA (2008). "Golgi appliance inheritance". The Golgi Apparatus: Land of the fine art 110 years afterwards Camillo Golgi's discovery. Berlin: Springer. p. 580. doi:10.1007/978-3-211-76310-0_34. ISBN978-3-211-76310-0.
- ^ a b c d Fabene PF, Bentivoglio 1000 (October 1998). "1898-1998: Camillo Golgi and "the Golgi": ane hundred years of terminological clones". Brain Research Bulletin. 47 (3): 195–8. doi:x.1016/S0361-9230(98)00079-iii. PMID 9865849.
- ^ Golgi C (1898). "Intorno alla struttura delle cellule nervose" (PDF). Bollettino della Società Medico-Chirurgica di Pavia. 13 (1): 316. Archived (PDF) from the original on 2018-04-07.
- ^ a b Davidson MW (2004-12-thirteen). "The Golgi Apparatus". Molecular Expressions. Florida State Academy. Archived from the original on 2006-xi-07. Retrieved 2010-09-20 .
- ^ a b c d eastward f g h Alberts, Bruce; et al. (1994). Molecular Biology of the Cell . Garland Publishing. ISBN978-0-8153-1619-0.
- ^ a b c d e Nakano A, Luini A (August 2010). "Passage through the Golgi". Current Opinion in Cell Biological science. 22 (4): 471–8. doi:10.1016/j.ceb.2010.05.003. PMID 20605430.
- ^ Suda Y, Nakano A (April 2012). "The yeast Golgi appliance". Traffic. 13 (iv): 505–10. doi:ten.1111/j.1600-0854.2011.01316.x. PMID 22132734.
- ^ Duran JM, Kinseth M, Bossard C, Rose DW, Polishchuk R, Wu CC, Yates J, Zimmerman T, Malhotra V (June 2008). "The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis". Molecular Biology of the Prison cell. xix (6): 2579–87. doi:10.1091/mbc.E07-ten-0998. PMC2397314. PMID 18385516.
- ^ Day, Kasey J.; Casler, Jason C.; Glick, Benjamin S. (2018). "Budding Yeast Has a Minimal Endomembrane Organization". Developmental Jail cell. 44 (i): 56–72.e4. doi:10.1016/j.devcel.2017.12.014. PMC5765772. PMID 29316441.
- ^ a b c d Day KJ, Staehelin LA, Glick BS (September 2013). "A three-stage model of Golgi structure and function". Histochemistry and Cell Biology. 140 (iii): 239–49. doi:10.1007/s00418-013-1128-3. PMC3779436. PMID 23881164.
- ^ Campbell, Neil A (1996). Biology (four ed.). Menlo Park, CA: Benjamin/Cummings. pp. 122, 123. ISBN978-0-8053-1957-vi.
- ^ William G. Flynne (2008). Biotechnology and Bioengineering. Nova Publishers. pp. 45–. ISBN978-one-60456-067-1 . Retrieved 13 November 2010.
- ^ Prydz K, Dalen KT (Jan 2000). "Synthesis and sorting of proteoglycans". Journal of Prison cell Science. 113. 113 Pt ii (2): 193–205. doi:10.1242/jcs.113.two.193. PMID 10633071.
- ^ a b c d e f g h i j g l m north o p q Glick BS, Luini A (November 2011). "Models for Golgi traffic: a disquisitional assessment". Common cold Spring Harbor Perspectives in Biology. iii (xi): a005215. doi:ten.1101/cshperspect.a005215. PMC3220355. PMID 21875986.
- ^ Wei JH, Seemann J (November 2010). "Unraveling the Golgi ribbon". Traffic. eleven (xi): 1391–400. doi:10.1111/j.1600-0854.2010.01114.10. PMC4221251. PMID 21040294.
- ^ a b Marie K, Sannerud R, Avsnes Dale H, Saraste J (September 2008). "Take the 'A' train: on fast tracks to the cell surface". Cellular and Molecular Life Sciences. 65 (18): 2859–74. doi:10.1007/s00018-008-8355-0. PMC7079782. PMID 18726174.
- ^ a b c d D'Souza-Schorey C, Chavrier P (May 2006). "ARF proteins: roles in membrane traffic and across". Nature Reviews. Molecular Cell Biology. 7 (5): 347–58. doi:10.1038/nrm1910. PMID 16633337. S2CID 19092867.
- ^ Papanikou E, Day KJ, Austin J, Glick BS (2015). "COPI selectively drives maturation of the early Golgi". eLife. 4. doi:10.7554/eLife.13232. PMC4758959. PMID 26709839.
External links
-
 Media related to Golgi apparatus at Wikimedia Eatables
Media related to Golgi apparatus at Wikimedia Eatables
Source: https://en.wikipedia.org/wiki/Golgi_apparatus
Posted by: placeneway1985.blogspot.com

0 Response to "Are Golgi Bodies In Plant And Animal Cells"
Post a Comment